I thought I'd share this video on RNA-Seq data analysis that I just watched. It was helpful to me so I thought it might be of interest to others as well. From here, I'm not exactly sure how to continue. Should I use the function on the overrepresented pvalues to correct for multiple testing?
RNA seq gives many genes showing differential gene expression, which could be due to some other factors as well but to narrow down for the exact candidate If you're more interested in overall patterns (GO analyses, how many genes diff. expressed), then don't do it. Also, if you have other types of
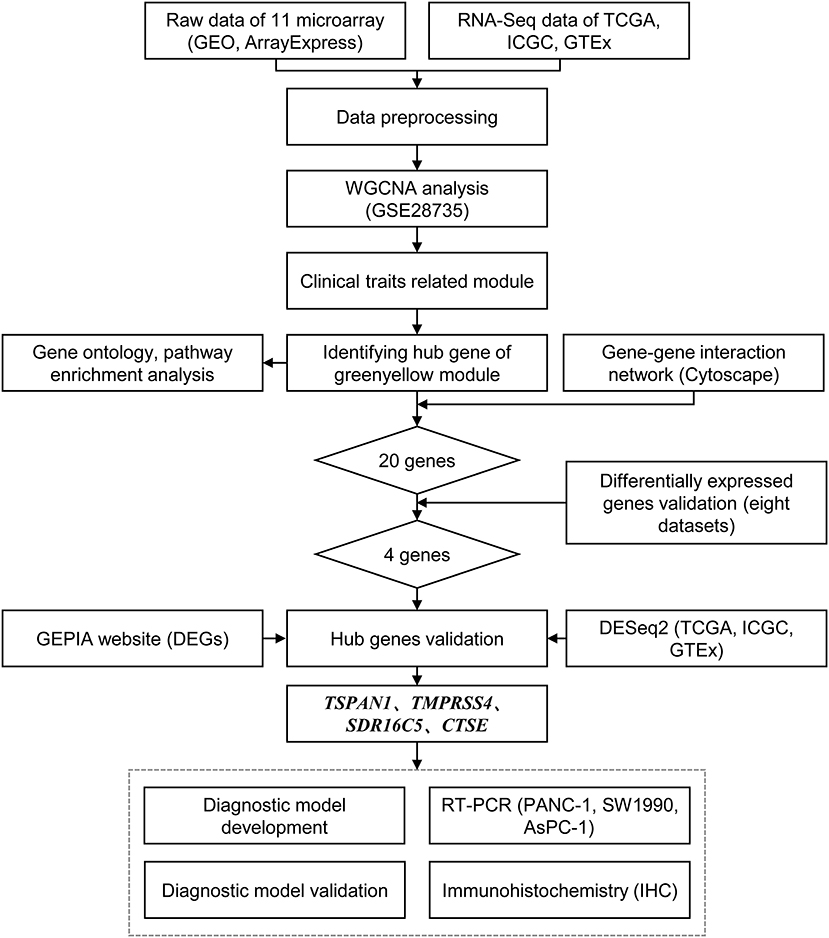
validation analysis frontiersin experiments pancreatic bioinformatics ctse novel cancer panel preparing flow development chart figure data
Genomic data are an excellent source of novel disease biomarkers. Single-cell RNA sequencing helps understand target expression. Genomic data are an excellent source of novel disease biomarkers and targets. In fact, genetically validated targets are twice as likely to achieve FDA approval (King et al.
NGS Validation, RNA-Seq, NanoString Single-Cell RNA-seq, Transcriptomics, Validation of RNA Sequencing. Researchers in the Spotlight--Download interviews to read how researchers use RNAscope ISH to validate RNA-seq results.
RNA-Seq (named as an abbreviation of RNA sequencing) is a sequencing technique which uses next-generation sequencing (NGS) to reveal the presence and quantity of RNA in a biological sample at a given moment, analyzing the continuously changing cellular transcriptome.
19, 2018 · RNA-seq is widely used for transcriptomic profiling, but the bioinformatics analysis of resultant data can be time-consuming and challenging, especially for biologists. We aim to streamline the bioinformatic analyses of gene-level data by developing a user-friendly, interactive web application for exploratory data analysis, differential expression, and pathway analysis. …
RNA-seq data analysis. The raw sequence reads from each replicate were aligned to the Ensembl human genome GRCh37 To validate data obtained from the RNA-seq analysis, we selected how to cite this article. Frost J, Ciulli A and Rocha S. RNA-seq analysis of PHD and VHL inhibitors

Validate RNA heterogeneity data Quantify protein heterogeneity Confirm gene expression results Correlate RNA-seq data to protein expression The following RNA-seq data validation information is presented in collaboration with Dhananjay Wagh, and John Coller, , Stanford

rna circrna cancer cell circular exome capture detection seq figure landscape validation method
18, 2022 · Yotsukura, S. et al. Celltree: An R/bioconductor package to infer the hierarchical structure of cell populations from single-cell RNA-seq data. BMC bioinformatics 17 , …
28, 2022 · I think that one of the questions is, indeed, the low sample size. We have four individuals in the case group and six controls. Regarding the expression of one gene of interest to the pathophisiology of the disease, we found these results in RNA-Seq: edgeR(log2FC = and p-value = ) and DESeq2 (log2FC = and padj = ).
How does RNA-seq work? RNA-seq vs microarrays: Why RNA-seq is considered superior An How does RNA-seq work? Early RNA-seq techniques used Sanger sequencing technology, a technique More quantifiable - Microarray data is only ever displayed as values relative to other signals
11, 2016 · Pertea et al. describe a protocol to analyze RNA-seq data using HISAT, StringTie and Ballgown (the ‘new Tuxedo’ package). The protocol …
While RNA sequencing (RNA‐seq) has become increasingly popular for transcriptome profiling, the analysis of the massive amount of data generated by large‐scale RNA‐seq still remains a challenge. RNA‐seq data analyses typically consist of (1) accurate mapping of millions of short
06, 2020 · For example, a recent study 30 used ribosome footprint profiling to validate 7,273 previously unannotated human genes, most of which were discovered when they ran StringTie on a very large set ( billion reads) of RNA-seq data. StringTie automatically detects new genes and new isoforms if they are present in your experimental data, regardless ...
RNA-seq RPKM Fisher's exact test Poisson LRT Negative Binomial. In this lecture we will begin to explore some of the issues surrounding more realistic models of dierential expression in RNAseq data. We will now examine how to perform an analysis for dierential expression on the basis of a
The data from scRNA-seq experiments are distinguished by their sparsity: in cell types where mRNA from ten Many computational methods have been proposed to re-duce the noise in single-cell RNA-seq data. We validate this approach on two datasets for which we have a form of ground-truth.
RNA-sequencing (RNA-seq) has a wide variety of applications, but no single analysis pipeline can be used in all cases. We review all of the major steps in RNA-seq data analysis, including experimental design, quality control, read alignment, quantification of gene and transcript levels,

aberrant ipf copd basaloid lungs fibrosis pulmonary advances idiopathic populations ectopic reveals
restriction map is a map of known restriction sites within a sequence of mapping requires the use of restriction molecular biology, restriction maps are used as a reference to engineer plasmids or other relatively short pieces of DNA, and sometimes for longer genomic are other ways of mapping features on DNA for longer length DNA …
The RNA-Seq data for the treated and the untreated samples can be compared to identify the effects of Pasilla gene depletion on gene expression. Data upload. In the first part of this tutorial we will use the files for 2 out of the 7 samples to demonstrate how to calculate read counts (a measure of the
Single-cell RNA sequencing (scRNA-seq) is a recent and powerful technology developed as an alternative to previously existing bulk RNA sequencing In addition, we consider a smaller subset of this data set consisting of solely the T-cell categories within the PBMC data set to better observe
04, 2011 · To validate the CIs generated by RSEM, we simulated an RNA-Seq data set with the mouse RefSeq annotation and estimated CIs with RSEM from 50% credibility up to 95% credibility. We then computed the fraction of transcripts for which the true abundances fell within the credibility intervals, out of all transcripts with abundance at least 1 TPM ...

qpcr fourteen validation
How does the SMARTer Stranded RNA-Seq Kit maintain strand information and why is this technique better than dUTP-incorporation methods? The SMART reaction is inherently stranded and does not require additional cDNA preparation steps to generate stranded RNA-seq data.
RNA-Seq has reached rapid maturity in data handling, QC (Quality Control) and downstream statistical analysis methods, taking substantial benefit from the extensive body of literature developed on the analysis of microarray technologies and their application to measuring gene expression.

identifies gene
The data we will be using are comparative transcriptomes of soybeans grown at either ambient or elevated O3 levels. Each condition was done in The Bench Scientist's Guide to statistical Analysis of RNA-Seq The samples we will be using are described by the following accession
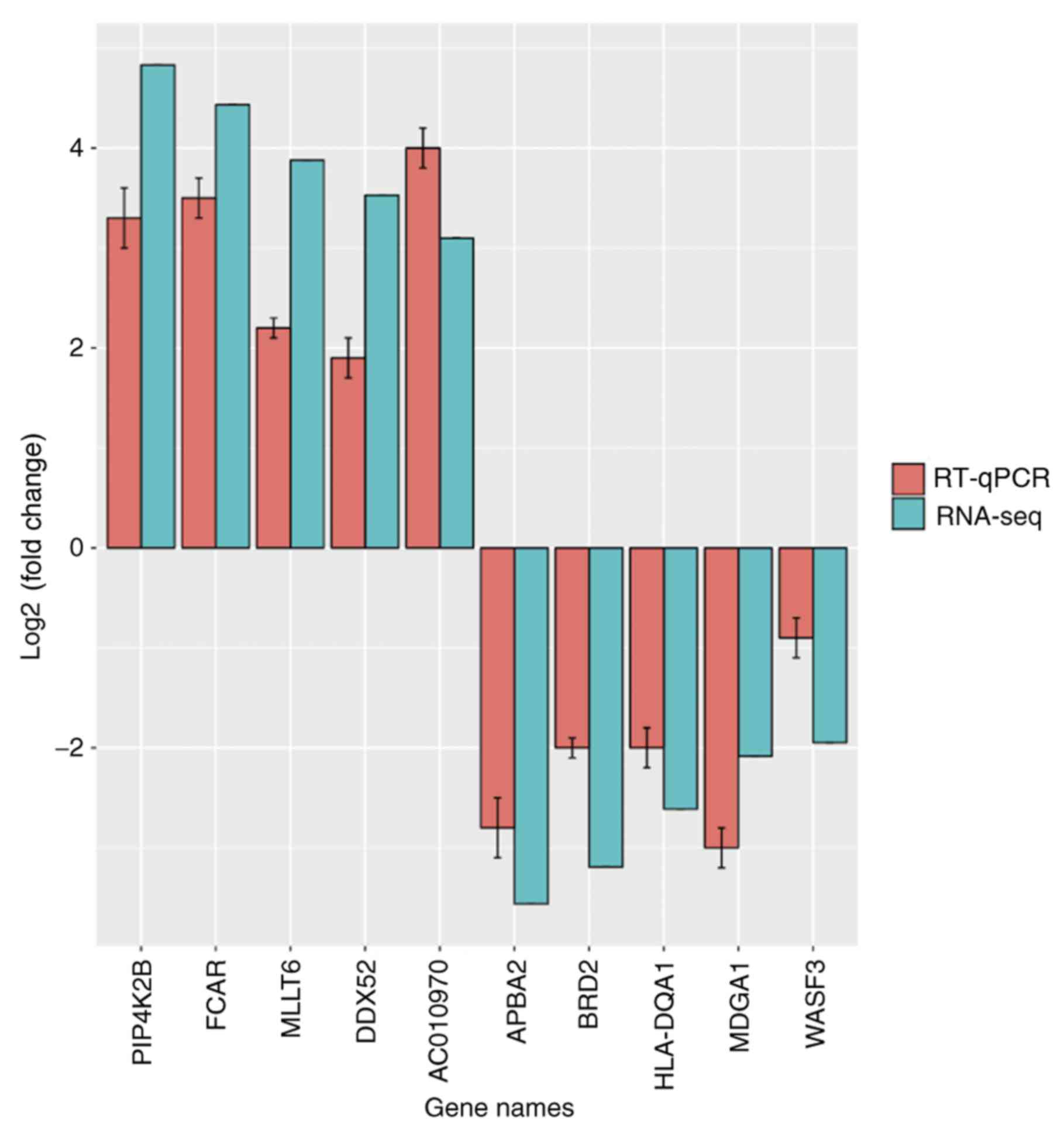
rna messenger acute syndrome distress respiratory qpcr seq neonatal sequencing mechanisms reveals between similar mmr
For those with RNA-Seq data, being asked for qPCR validation is often met with surprise. Let's take a look at situations when qPCR validation of If you plan to validate your RNA-Seq results using qPCR, then we highly recommend using a different set of samples with proper biological replication.
11, 2019 · We introduce APEX-seq, a method for RNA sequencing based on direct proximity labeling of RNA using the peroxidase enzyme APEX2. APEX-seq in nine distinct subcellular locales produced a nanometer-resolution spatial map of the human transcriptome as a resource, revealing extensive patterns of localization for diverse RNA classes and transcript isoforms.
If you benefit from my tutorial and use the same strategy for data analysis, please CITE my RNA-Seq paper published in "Scientific Reports - Nature":
RNA-seq data analysis. Posted on September 13, 2016. Below shows a general workflow for carrying out a RNA-Seq experiment. In this guide, I will focus on the pre-processing of NGS raw reads, mapping, quantification and identification of differentially expressed genes and transcripts.
Informatics for RNA-seq: A web resource for analysis on the cloud. Educational tutorials and working pipelines for RNA-seq analysis including an introduction to: cloud computing, critical file formats, reference genomes, gene annotation, expression, differential expression, alternative splicing,
31, 2022 · All data generated or analysed during this study are included in the manuscript and supporting file; Source Data files have been provided for all figures. ... of replicative senescence in WI-38 human lung fibroblasts equipped with a battery of modern techniques including RNA-seq, single cell RNA-seq, proteomics, metabolomics, and ATAC-seq ...
GSEA requires as input an expression dataset, which contains expression profiles for multiple samples. While the software supports multiple input file formats for these datasets, the tab-delimited GCT format is the most common.
10, 2020 · A range of techniques such as FISH, immunohistochemistry (IHC), and RNA sequencing (RNA-seq) were used to yield a comprehensive database—eMouseAtlas data (Armit et al., 2017), which can be utilized to validate new spatial technology. We applied DBiT-seq to an E10 whole mouse embryo tissue slide at a pixel size of 50 μm to computationally ...
A standardized pipeline for RNA-seq analysis eliminates data processing variation. The GeneLab RNA-seq pipeline includes QC, trimming, mapping There is presently no consensus on how best to analyze RNA-seq data, and the impact of analysis tool se-lection on results is an active eld of research.
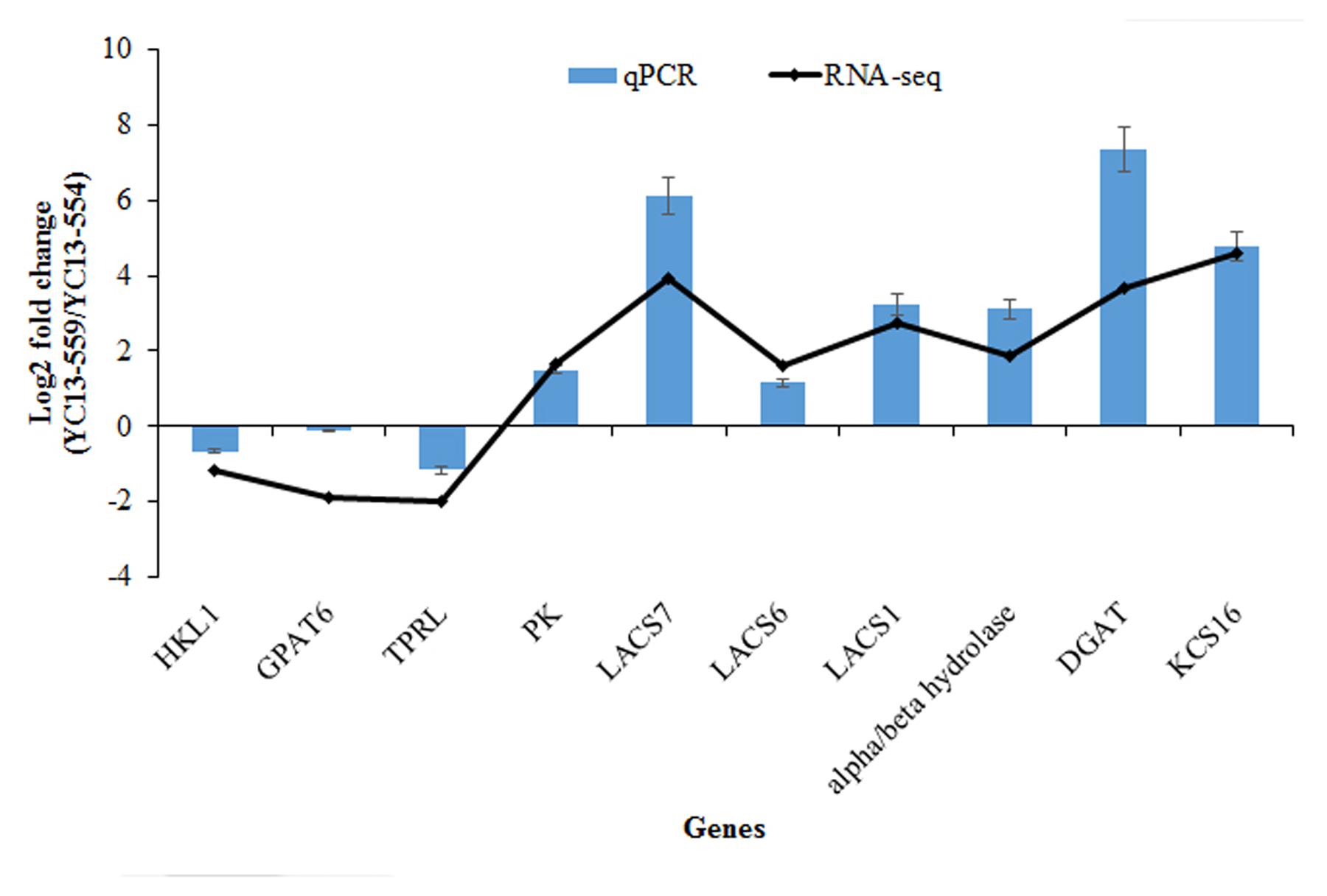
plant lines analysis brassica napus frontiersin rna seq genes accumulation comparative isogenic transcriptomic influences reveals seed network oil near fpls
Bulk RNA-seq. A major breakthrough (replaced microarrays) in the late 00's and has been This course is concerned with the computational analysis of the data obtained from scRNA-seq The strategy used for capture determines throughput, how the cells can be selected as well as what
Additional information on how RNA extraction methods inuence RNA-seq data can be found in Sultan et al. If you are interested in performing power analyses for dierential gene expression detection using RNA-seq data, you can have a look at the publication and R code provided by Ching et al.

transcriptome copd seq inflammation asthma
RNA-seq technologies enable the measurement and comparison of genome-wide gene Meanwhile, no matter how complicated a statistical model is, its general framework and logical Experimentally validated benchmark data for RNA-seq experiments are still limited on the genome-wide scale.
The RNA-seq workflow describes multiple techniques for preparing such count matrices. It is important to provide count matrices as input for DESeq2's A tutorial on how to use the Salmon software for quantifying transcript abundance can be found here. We recommend using the --gcBias flag
Can anyone tell me how to validate the RNA-Seq Differential gene expression data using qPCR? RNA-Seq relay in the single fact that a sequence read, as long as 100 or 150 bases is undoubtedly mapped to our genome or transcriptome.
Data Availability: RNA-seq data have been deposited at the European Genome-phenome Archive on simulated and real RNA-seq data, and illustrate how four different read aligners affect prediction results. We validate our approach for quantification based on simulated and real RNA-seq
RNA-seq generates vast amounts of data and when all of this is sorted out and boiled down to results, the researcher is usually left Right now, there are no standard guidelines for when or exactly how RNA-seq data should be validated. So let's look at some of the arguments for and against validation.
