This video explains how to use Nanodrop to measure and access quality of DNA/RNA following extraction. Please ask me any ... NANODROP is a full spectrum (220nm-720nm) spectrophotometer and can be used to measure concentration of Proteins ...
concentration using the Bradford assay • Protein Lowry - protein concentration using the The user also has options as to how the buffer is handled. Refer to section 14 (Data Viewer) for Measurement Concentration Range The NanoDrop® ND-1000 Spectrophotometer will
Calculations of protein concentration from absorbance values were automatically performed by the NanoDrop 2000c software. Absorbance data was exported from the reference spectrophotometer and protein concentrations manually calculated according to Beer's Law using an E1% of

labeling antibody igg

protein purification proteins expression imidazole igem purdue purified
Quantifying a DNA, RNA or protein sample concentration is now as easy as a click of the pipette, a push of a button and a dab of tissue to clean up. A Nanodrop is a common lab spectrophotometer (you may already be familiar with the 1000 or 2000 model) that reads a single 2?l drop on a pedestal.
The NanoDrop Lite should be used to measure the concentration of purified protein samples. Q: What is the dynamic (concentration) range and reproducibility for proteins on the. NanoDrop Lite? A: The dynamic range depends on the assay type selected (1A/cm = 1 mg/ml, IgG or BSA).
I've been using nanodrop for a long time, however I ised it only for RNA/DNA concentration analysis. In fact, there's a "protein" option in nanodrop that I have never used before, though one of my colleagues recently noted that they use it frequently. Actually what I heard about protein measure
Microvolume Protein Concentration Determination using the NanoDrop 2000c Spectrophotometer. This video explains how to use Nanodrop to measure and access quality of DNA/RNA following extraction. Please ask me any ...
...NanoDrop Eight and NanoDrop One instruments calculated the corrected dsDNA concentrations with phenol and protein contamination. The channel-to-channel variability for the NanoDrop Eight Spectrophotometer was also evaluated using the 3 mg/ml spike sample for protein and the

nanodrop thermo 2000 spectrophotometer scientific 2000c spectrophotometers dna rna uv equipment protein vis purity thermofisher fisher nd 1000 spectroscopy spectrophotometry
BSA Protein concentrations are calculated using the mass extinction coefficient of at 280 nm for a 1% (, 10 mg/mL) FAQs Q: How do I calibrate NanoDrop spectrophotometers? A: The Calibration Check diagnostic allows the user to confirm that the instrument is performing within specifications.
For proteins suspended in a RIPA buffer, it is recommended protein concentration be determined using a RIPA buffer compatible colorimetric assay. Desjardins, P., Hansen, J. B., Allen, M. Microvolume Protein Concentration Determination using the NanoDrop 2000c Spectrophotometer.
The obvious measurement that the Nanodrop produces is the concentration of the sample in ng/uL. The main reason people use the Nanodrop is to deduce the purity of their samples. The 260/280 ratio gives an indication of how pure the sample is from contaminating protein.
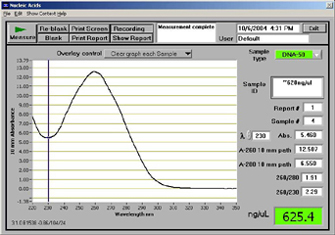
nanodrop spectrophotometer dna measurement output samples second core
Using the NanoDrop 2000c for Protein Crystallography. Microvolume Protein Concentration Determination using the NanoDrop 2000c SpectrophotometerПодробнее. DNA quality and quantity measurement by NanoDrop 2000c SpectrophotometerПодробнее.
Visit for more information about NanoDrop Thermo Scientific NanoDrop 2000c Spectrophotometer employs an innovative

nanodrop

Page 66 Calculates lysozyme protein concentration using mass extinction coefficient ( ) of Page 94 Protein assay kits and protocols Please refer to the NanoDrop website for up-to-date kits and Page 96 The R value indicates how well the standard curve fits the standard data points (
...Concentration Using Nanodrop The Nanodrop is shared equipment among research and teaching labs, so be mindful of other users' time, and only use Materials: · Protein samples · Nanodrop device · Container with ice · 10mL micropipette with tips · Kimwipes · Vortex mixer * ·

Calculations for Protein A280 Measurements. Measure Proteins and Labels. Can be used to correct for any offset caused by light scattering particulates by subtracting measured absorbance at specified baseline correction wavelength from absorbance values at all wavelengths in
I know how to use the Nanodrop, but when I get the spectra and all the numbers, I'm not really sure what to do next. I know absorbance is proportional to If I'm trying to find the concentration of protein only (I'm trying to remove RNA actually, so I am hoping I disassembled the virus and purified
How does the Nanodrop convert A260 to concentration in ng/uL? For MicroArray, UV-Vis, Protein BCA, Protein Bradford, Protein Lowry and Cell Culture modules the data are normalized to a cm ( mm) path.
To determine antibody concentration using a NanoDrop spectrophotometer, follow these steps (the exact procedure may vary based on your NanoDrop model): 8. Place a 2 µL aliquot of antibody solution on the clean pedestal and click the 'Measure' icon.
Thermo Fisher NanoDrop™ technologies perform protein concentration measurements using a pedestal measurement technique. A sample droplet is pipetted directly onto the spectrophotometer's lower pedestal, a column is formed when the upper surface is brought in contact with the sample

nanodrop spectrophotometer igem newcastle
Nanodrop® technology can be used to measure the concentration of individual synthetic oligos using each oligonucleotide's unique analysis constant. These numbers are used to calculate the analysis constant needed for Nanodrop concentration calculations.
Concentration units may be selected from the adjacent drop-down box. Beer's law is used to calculate the nucleic acid concentration as described in 'Nucleic Acid Calculations'. This method does not require generation of a standard curve and is ready for protein sample quantitation at software startup.
Learn how to accurately determine the concentration of your oligonucleotides using Thermo Scientific NanoDrop instruments. The default setting for NanoDrop instrument measurements is 340 nm. This correction is necessary, because it adjusts for any light scattering event that may skew results.
NanoDrop is a Spectrophotometer that measure 2 ml samples with high accuracy and reproducibility. The NanoDrop model offers the convenience of both the NanoDrop patented sample retention technology and a traditional cuvette for sample measurements.
Microvolume Protein Concentration Determination using the NanoDrop 2000c Spectrophotometer. Thermo Scientific NanoDrop UV-Vis spectrophotometers support protein sample quantification with applications for direct A280, A205 and colorimetric assays .
Both Rutger's University and University of Toronto use the UV absorbance at 280 nm to determine protein concentrations. Absorbance by proteins is primarily due to the aromatic rings of Phe, Tyr, and Trp residues.
A review of the NanoDrop for Measuring Nucleotide and Protein Concentrations . Unbiased reviews by scientists available at This is a must-have instrument for many biology labs where measuring accurate DNA or RNA concentration is essential for daily experiments.

nanodrop thermo spectrophotometer lite scientific fisher 120v spectrophotometers
Interpreting Nanodrop (Spectrophotometric) Results. Foundation of Spectrophotometry: The Thus, the amount of light absorbed in this region can be used to determine the concentration of A260/280 ratio The A260/280 ratio is generally used to determine protein contamination of a nucleic acid sample.
In addition to concentration, protein measurements on the NanoDrop One spectrophotometer deliver information about contaminants in the sample. The NanoDrop One Thermo Scientific™ Acclaro™ Sample Intelligence Technology uses mathematical algorithms to detect nucleic acids in
Use of the Nanodrop Spectrophotometer. Open the software of the Nanodrop by double clicking at the icon "ND-1000 " on the desktop. ¾ To measure the protein concentration at 280nm press the "Protein A280" button.
