
methods

crispr cas9 rna guide sequence step system cas grna flies cartoon editing genome target gene parts relative schematic zebrafish loxp
Gene Link shRNA Design Guidelines. GeneID: Accession Number shRNA construct options. Loop Options TTCG TCAAGAG GAAGCTTG other. RNA Length 23 mer 29 mer other.
BackgroundShort hairpin RNA (shRNA) encoded within an expression vector has proven an effective means of harnessing the RNA interference (RNAi) pathway in @article{Mcintyre2006DesignAC, title={Design and cloning strategies for constructing shRNA expression vectors}, author={Glen
Versus siRNAEffective Expression of shRNAs from RNA Polymerase III PromotersLentiviral Vectors Enable Permanent Knockdown of TargetsOptimization of shRNA DesignClick here for the PDF version RNA interference (RNAi) is a biological process where RNA molecules are used to inhibit gene expression. Typically, short RNA molecules are created that are complementary to endogenous mRNA and when introduced into cells, bind to the target mRNA. Binding of the short RNA molecul…See more on
(Upper) The general vector design with the two independent shRNA and TuD expression cassettes, together with selected unique restriction sites and Because reporter knockdown assays do not necessarily reflect how shRNAs silence their genuine targets, we next examined the effects of
What is shRNA and how do you use it? Short hairpin RNA (shRNA) sequences are usually encoded in a DNA vector that can be introduced into cells shRNA technologies are DNA-based, which provides flexibility in design of the vector. Most vector-based shRNA systems contain a selectable marker
This lecture is about shrna design and knockdown mechanism of gene using shrna mediated gene silencing. How do recombinant lentivirus systems work? Lentiviruses are members of the Retroviridae family of viruses, with HIV-1 being the ...
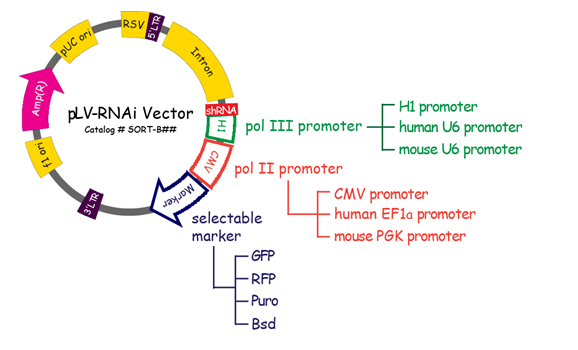
rnai plv shrna transduction lenti lentiviral protocols analysis
Optimization of shRNA Design. Designing effective shRNAs is essential for effective RNAi knockdown screening. To efficiently test shRNA structure variations on a large-scale, we developed a high-throughput shRNA efficacy testing technology based on a reporter assay.
We design shRNA constructs ("clones") with an algorithm. Our algorithm uses several criteria to rank potential 21mer targets within each human and mouse Refseq transcript. The algorithm applies a set of rules, including those derived from the siRNA literature, analysis of TRC library performance
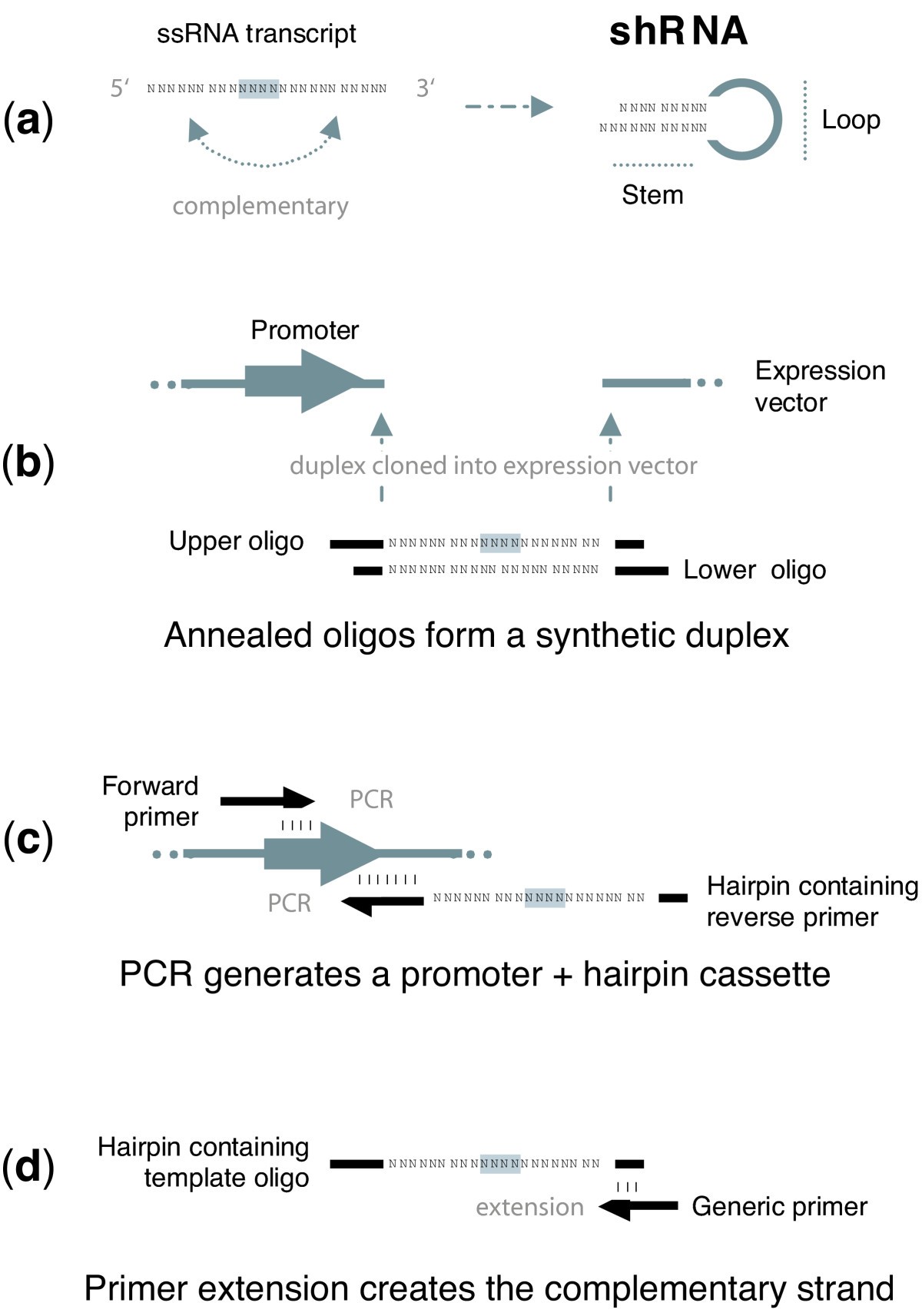
shrna expression vectors cloning constructing strategies figure rna constructed different
How is shRNA knockdown score calculated? VectorBuilder applies rules similar to that used by the RNAi consortium (TRC) to design and score shRNAs. For each given RefSeq transcript, we search for all possible 21mers that are considered as candidate target sites.
Using a specially designed expression vector, the shRNA can be synthesized in vivo ( in a cell). Expression vectors are a great option to elevate the artificial shRNA-mediated process. If you know how vectors work and manufactured, perhaps; you can understand how the process is done.
Currently, major shRNA/siRNA design algorithms have focused on knock-down efficacy over specificity. Here, we present a novel ap-proach to design potent shRNA/siRNA with minimal off-target effects. 11. Gu,S. and Kay, (2010) How do miRNAs mediate translational repression?
We also provide a real example demonstrating how to keep track of those variances in order to estimate confidence intervals (C. I.) at the 95% level. Using this theoretical error model and our standardized experimental protocol, we have tested 329 SureSilencing™ pre-designed
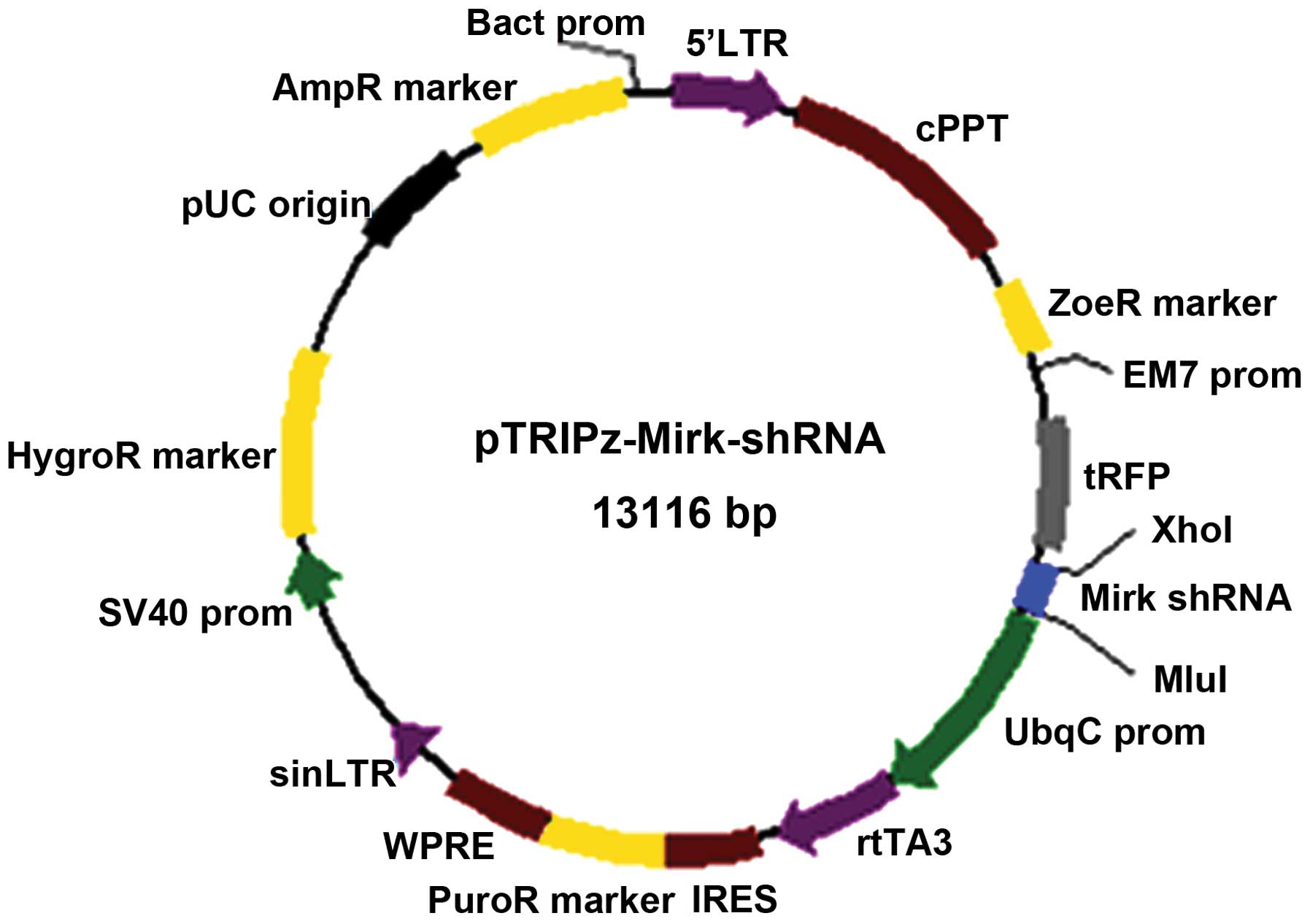
lentiviral tet ires cppt shrna vector lentivirus construction vectors rfp mirk ltr rnai gene element mediated fluorescence ijo human purine
How can you remove a single ruler guide while leaving others in place?
How do I screen arrayed shRNA libraries? Where can antibodies against puromycin can be purchased? What are the advantages of selection When conducting experiments using MISSION® TRC shRNA constructs, the proper controls should be a key element of your experimental design
Hi everyone, I would like to create a pool of shRNA sequences (20nts each) for the specific set of 1000 genes, making it up to 30000 sequences in total, the gene sequences are available There are online tools that design shRNAs for single genes, but I can't figure out how to do this for 1000 sequences.
How I design a T Shirt - Clothing Art Tutorial Ten Hundred Ten
the design of shRNA that will be expressed from a U6 or 7S K promoter, select target sequences beginning with a G. Perform a BLAST search to eliminate potential target sequences that have a perfect match of 16 nt or more to an off-target gene of the same species ( ).Author: Chris B Moore, Elizabeth H Guthrie, Max Tze-Han Huang, Debra J TaxmanPublish Year: 2010
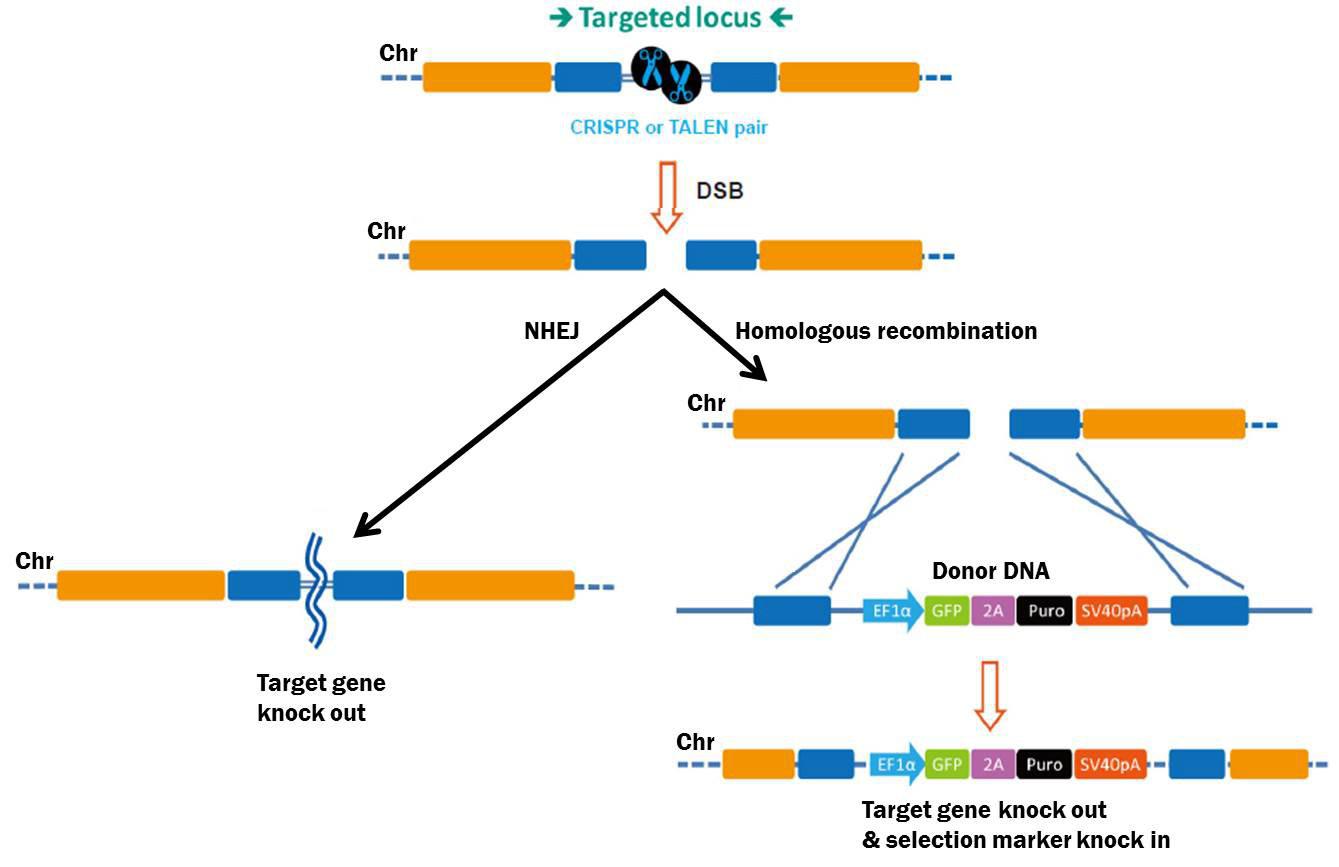
talen crispr knockout genome knockdown donor sirna dsbs homologous recombination nhej joining sgrna shrna rna
Practical advice for using CRISPRs: designing your gRNA and introducing it into cells. While this seems obvious, it is important to remember that the same is true when designing gRNAs for using CRISPR technology - the "best" gRNA depends an awful lot on what you are trying to do:
The design of siRNAs and short hairpin siRNAs (shRNAs) remains an empirical process since the molecular mechanisms underlying RNAi are not yet Several teams including ours have tested a variety of sequences for the loop between the two complementary regions of a shRNA, ranging
how_to_design_an_shRNA. Dr. Guihua Sun (Rossi Group) Glackin Group Target sequence siRNA 5B-Tw418,GATCGGACAAGCTGAGCAAGATTCATTTGTGTAGTGAATCTTGCTCAGCTTGTCCTTTTT Seed seq Clone U6-shRNA fragment into pENTR shuttle vector to be delivered to pBLOCKiT6.
How it works in theory is rather simple: Let's take the Bored Ape Yacht Club as an example. An ape is composed of different properties such as background Here is the good news though: In this guide I will not only show you step by step how you can create your very own collection, without you
Four shRNA constructs were designed to target Kat60 in a species-specific manner. Exon number and species name indicates which exon and allele of Kat60 should be designed to have mismatches with likely off-targets as close to the PAM as possible. Off-target effects can be further reduced by
Design And Cloning Strategies For Constructing Shrna Expression. Dazl Shrna Construct Design Dna Oligonucleotides Containing Dazl. Multiplex Shrna Screening Of Germ Cell Development By In Vivo. Shrna Knockdown Shrna Clones Lentiviral Shrna Genecopoeia.
The RNAi Designer incorporates the guidelines provided in this manual as well as other design rules into a proprietary algorithm to design shRNA When designing your RNAi experiment, you should consider how to assay for knockdown of the target gene. After you have transfected your
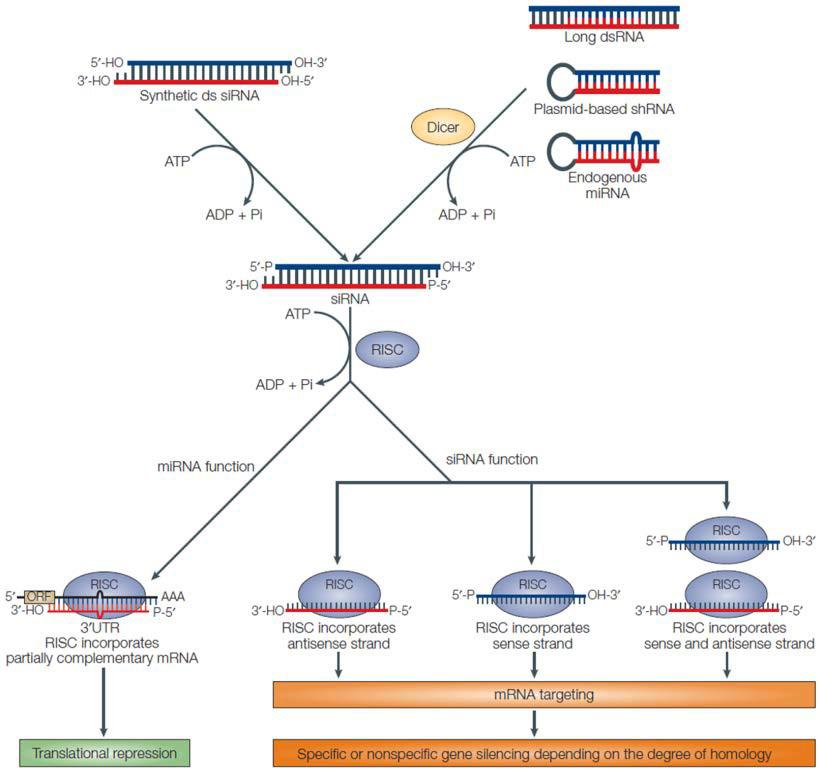
talen knockout rnai crispr sirna shrna vs knockdown scheme pathways general illustration genecopoeia mittal 2004 figure
Short hairpin RNA (shRNA) encoded within an expression vector has proven an effective means of harnessing the RNA interference (RNAi) pathway in mammalian cells. In this study we detail simple improvements for constructing and sequencing shRNA that overcome current limitations.
How to design shRNA? any suggestions? tools or software to use for this designing? I am currently designing oligo sequences for depletion by shRNA. I have found a lot of on line tools (below) but each one give me different oligo sequences and I just do not know which I should use.

shrna addgene plko cloning rna hairpin trc japaneseclass

lenti gfp shrna cas9 bdnf geg crispr cre tech vectors myod nls lentivirus vector lentiviral rna expression
) Design at least 3 shRNAs per gene. All of the design principles for siRNAs apply. In addition, you can pick oligos that can work for both mouse and human genes (this has indeed been done for the library at CSHL), allowing use of common oligos in both species. 2) Barcodes (random 60 mer sequences synthesized in a lab) are inserted into the Size: 63KBPage Count: 3
9. SureSilencing shRNA - How it works A vector is introduced into cells and utilizes the U1 promoter to ensure that the shRNA is always expressed Dicer cleaves 29. shRNA Case Study #1- Experimental Workflow Validation of STAT3 RNAi Design Total RNA->cDNA Reverse Transcription:
siRNA/shRNA/Oligo Optimal Design. How. Details: siRNA/shRNA/Oligo Optimal Design. shRNA (short hairpin RNAs) are artificial constructs that can be expressed endogenously.
AGAUGUA CAC Antisense sequence of shrna shrna mir design details 1,2,3 Replaced mature 4 GUARANTEED KNOCKDOWN WITH EXPRESSION ARREST shrna mir OPEN BIOSYSTEMS INTEGRATED DNA TECHNOLOGIES, INC. Dicer Substrate RNAi Design How to design and
have made repeated attempts to clone shRNA into pLKO vectors, but have not produced any colonies. I have tried annealing the shRNA in a thermocycler, in PCR, and in a water Reading Time: 4 mins
All shRNA were designed to produce potential siRNA-like cleavage product with the optimal How can we use duplex stability of fully paired siRNA or shRNA antisense strands (further, duplex Software for efficient siRNA and shRNA design which employ application of optimal thresholds
Reliable design of these molecules is essential for the needs of large functional genomics projects. We identified features of the shRNA guide strand that significantly correlate with the silencing We used Sn stability value as an indicator of how much the parameter's predictive power depends on
