Primer design is like art. There is more than one design to cover the region of interest. Are you an "Artist"? Primers are crucial to the success of target amplification and subsequent sequencing in PCR and Sanger sequencing workflows.
This video shows how to design primers for PCR (and probe for Real time quantitative PCR).
My PI suggested me to design internal COI primers for Peringia ulvae. How can I do that? Primers generally give good reads from the 5' end for about 600-700 bp and after that the quality of base calling deteriorates, so if your target amplicon is above 1500bp or so, you would need internal

circrna pcr circular rt biogenesis schematic divergent sequence junction mrna analysis pre rnas amplification detection protocol primers template representation figure
Design a set of sequencing primers from EST sequences that as markers are associated with some BAC clones and contigs of D genome physical maps. A negative value is assigned counting. from the end of the sequence. This tells you how. far a primer is picked from 3' UTR.
Miscellaneous Sequencing Primers. PinPoint™ Sequencing Primer 5′-d(CGTGACGCGGTGCAGGGCG)-3′ pTargeT™ Sequencing Primer 5′-d(TTACGCCAAGTTATTTAGGTGACA)-3′. V4211 Q4461.
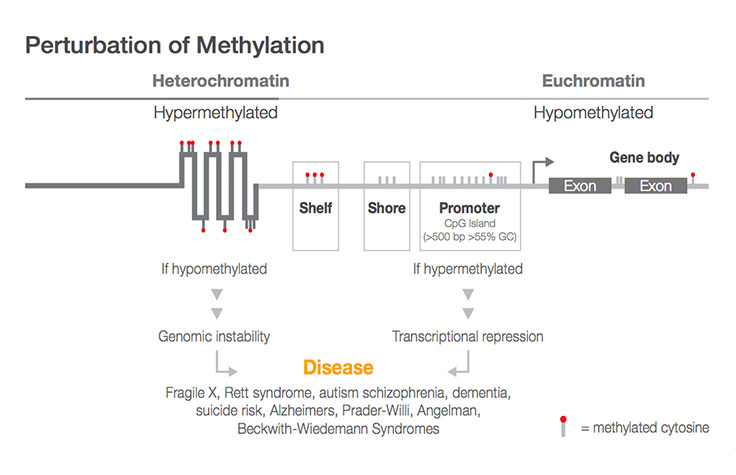
methylation array sequencing illumina analysis genome whole data dna bisulfite pcr seq rna sequence techniques marketing tips coverage
Customer data for Guide-it products. How to design sgRNA sequences. Introduction to the CRISPR/Cas9 system. Gene editing of CD3+ T cells Using the most effective sgRNA. Even if you pick a target sequence that fulfills all of the described requirements, sgRNA specificity and activity
Sequencing primers which have been designed to bind to our region of known DNA with the 5' to 3' orientation pointing towards your region to be Sequencing primers are designed using the same criteria we would use for PCR primers. Identify appropriate sequencing primers from empty vector.
How do I design primers for sequencing? We recommend designing your sequencing primers in a region that is 100 bases upstream of your sequence of interest. If you do not have the luxury of having this buffer, the closest you want the primer to be to your area of interest is 50-60 bases.
I have designed custom sequencing primers to be in the ranges of the standard Illumina sequencing primers: length = 33bp, GC content = and Tm = ºC (from OligoAnalyzer). ZAAB, suggested that the Tm should be higher than
Reverse primer regions where a sequencing primer can start. These are defined by making a selection on the sequence and right-clicking the selection. If areas are known where primers must not bind ( repeat rich areas), one or more. No primers here regions can be defined.
Design forward and reverse sequencing primers using our free PrimerQuest Tool . Primers should be between 18-30 nt long with an optimal length of 20-25 nt. They should have a G-C content between 40-60% and a T m between 55-65°C. Avoid primers that form strong self-dimers or hairpins,
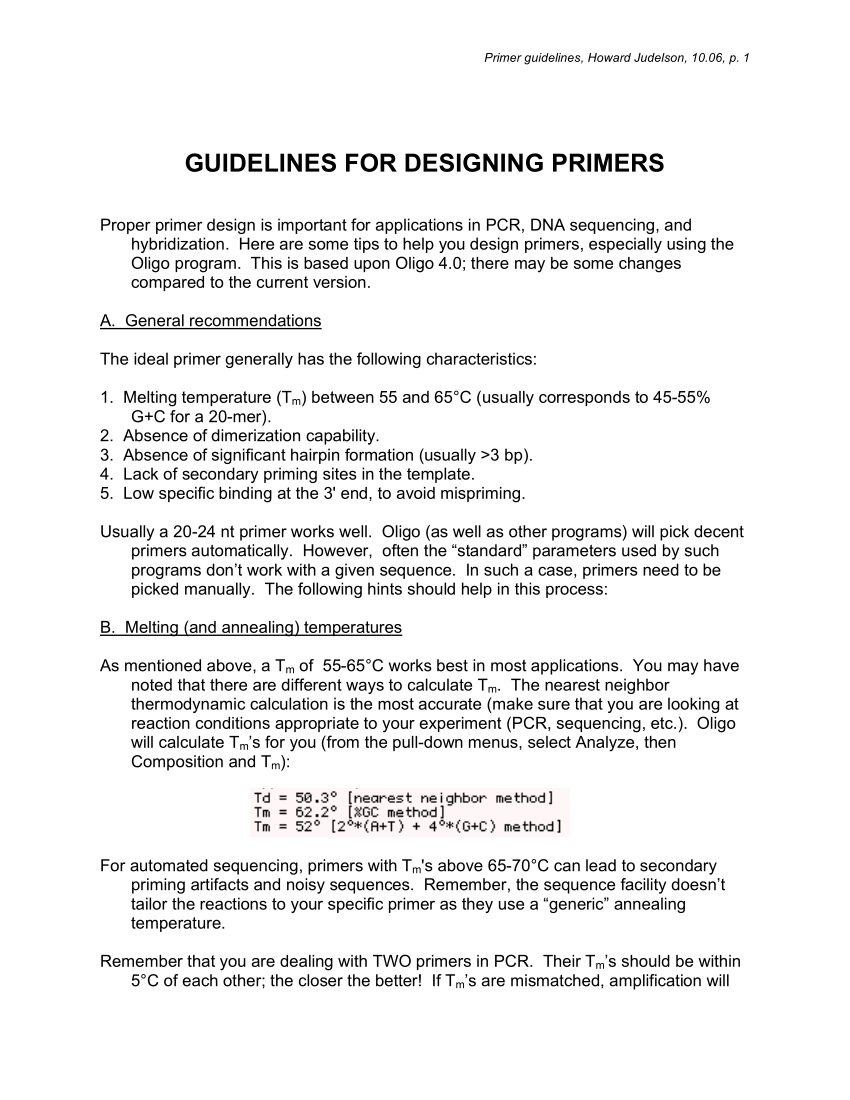
Primer Design for PCR: Primer guidelines page offers a look at the general and useful guidelines laid for designing primers for PCR reaction The primers are then annealed to the complementary regions of the single stranded molecules. In the third step, they are extended by the action of the
Enter one or both primer sequences in the Primer Parameters section of the form. If only one primer is available, a template sequence is also required. If the NCBI mRNA reference sequence accession number is used, the tool will automatically design primers that are specific to that splice variant.
I did what most people seems to suggest: Plug the standard primer into the IDT Tm calculator (66C), then design a custom primer that is similar in Keep in mind that the conditions of flowcell annealing and "sequencing-by-synthesis" are quite different than those in your thermal cycler (
Proper primer design is one of the single most important factors in successful automated Sanger DNA Sequencing. The following criteria are considered most critical in sequencing primer design: Primer length should be in the range of 18 and 24 bases.
Elegant primer design tool. Design PCR and sequencing primers and hybridization probes, to any target region or entire sequence, directly on alignments and assemblies in the Geneious sequence viewer.

sequencing pcr sanger primers thermofisher workflow

oligonucleotide degenerate primers cloning restriction sequencing nucleotide ndei bamhi
Primers designed according to these criteria will generally be from 18 to 30 bases in length and have %GC of 40 to 60. Try to avoid using primers with Tm's above 65-70 C, especially on high GC templates, as this can lead to secondary priming artifacts and noisy sequences.
Terminal nucleotides in the primers. How to design PCR primers? A step-wise guide DNA primers are easy to synthesize and use in comparison to RNA primers. We are amplifying DNA not RNA so ideally it's recommended to use DNA primers.

allele genotyping validation promoter mgmt

primer dimer primers hairpin dimers example designing analysis thoughts sequences genetics
primers designing guidelines
GenScript DNA Sequencing Primers Design Tool. ** This online primer design tool helps you to design primers for sequencing. You can customize the approximate distance between sequencing primers and the Tm (melting temperature) range.
How do you design primers for sequencing plasmids? The primer based approach was used with Sanger sequencing (or primer extension in the presence of chain terminators). This approach is too slow and cumbersome.

sequencing amplicon clinical
How extension products are labeled. Automated cycle sequencing procedures incorporate fluorescent dye labels using either dye-labeled With dye primer chemistry, four separate tubes of sequencing primers are each tagged with a different fluorescent dye. 2. Design primers. 3. Clean up templates.
Here's how to proceed: I. Design primers only from accurate sequence data. Automated sequencing (and in fact any sequencing) has a finite probability For a sequence assembly project, design more primers than you think you really need so that if the sequence isn't as long as you hoped, you
Eurofins Genomics' Sequencing Primer Design Tool uses Prime+ of the GCG Wisconsin Package originally written by Irv Edelman. With the sequence primer design tool, you can design forward and reverse sequencing primers for a target of interest. The optimal parameters for a primer
Would the sequencing primers just be the primers I used for PCR? If not, how would I design a sequencing primer for my PCR fragments? Primers with long runs of a single base should generally be avoided. It is essential to avoid 3 or more G's or C's in a row.
Ever designed sequencing primers before that didn't work? It can be extremely frustrating, right? Trust me, I know exactly how you feel. But you definitely don't want to design sequencing primers that ultimately fail. That can be days or a couple of weeks wasted because you're waiting for
GenScript Sequencing Primer Design. How. Details: GenScript DNA Sequencing Primers Design Tool ** This online primer design tool helps you to design primers for sequencing.
Eurofins Genomics' Sequencing Primer Design Tool is using Prime+ of the GCG Wisconsin Package originally written by Irv Edelman. The design tool analyses the entered DNA sequence and chooses the optimum forward or reverse sequencing primers. In selecting the appropriate primers,
of the Illumina sequencing primers, the addition and mixing of custom primers, it is recommended to use slimmer tips or larger volumes of lower stock If only the custom primer is needed, custom primers can be used in place of the Illumina primers. For instructions on how to set up a run

primer primers oligo tools designing thermofisher tips nti vector software thermo fisher dna sequence analysis steps invitrogen
How to Design a Primer. Diagnostic Restriction Digest. These primers typically anneal to the backbone and can help you verify the ends of the insert. If you require additional sequencing and need to design a custom primer, Addgene recommends using Addgene's sequencing results as
Primer Design, 16S Amplification and Sequencing Procedures. Primer design for universal amplification of the V4 region of 16S rDNA was based on a protocol published by Caporaso and Our primer sequences and staggered sequencing strategy are described in detail in the
