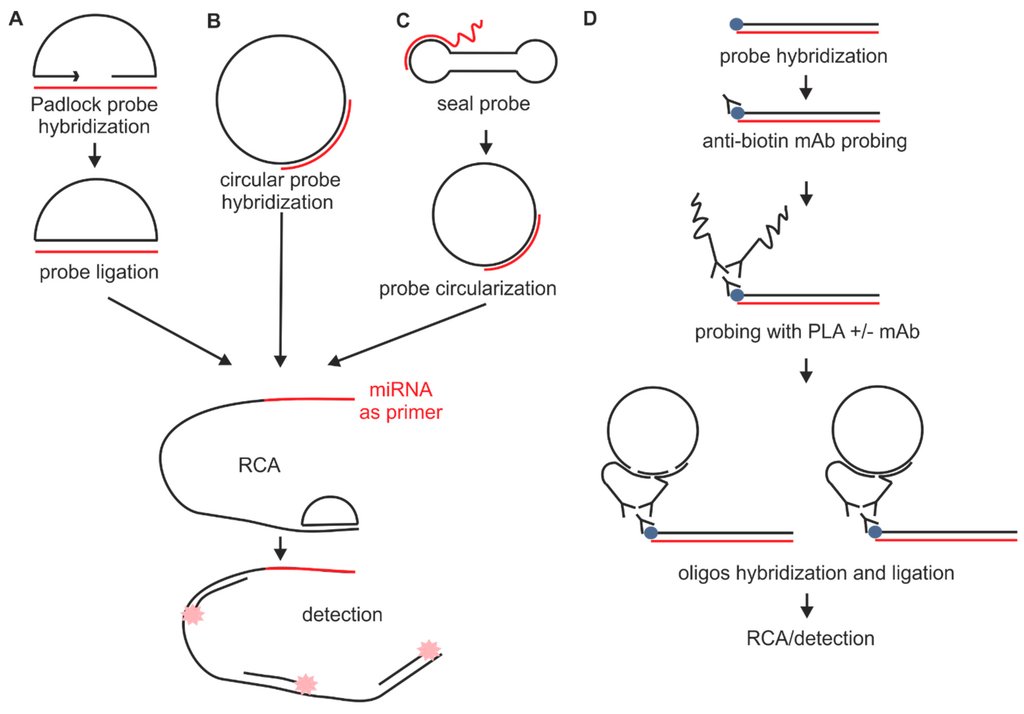
hybridization rna situ probes detection dna mirna amplification circular sequence ish ligation methods ijms target ligase mdpi molecules g003a
Design Group-Specific FISH Probes. Probes are designed to maximize sensitivity and specificity to the target group(s) (identifier(s)). If numProbeSets > 0 then that many pairs of probes with minimal cross-hybridization overlap are returned, enabling increased specificity with a dual-color approach.
How to design a FISH probe? - ResearchGate. Windows. Details: Asked 23rd Jul, 2015. Sivarajan Chettiar. Hello, I want to design FISH probes You may use the designer for in situ probe design, review your sequences, save them to your cart, and proceed to online checkout with just a

fish probes fixation protocol cell principle card protocols fluorescently standard arb silva

ozone generator ras mat controls fish

fish probe stellaris rna biocompare company reagent
Do you provide design assistance for FISH probes? We offer complementary FISH probe design service using our proprietary design algorithm for How many FISH assays can you get from 500pmol of labeled probes? The number of assays per library depends on a number of factors including
Centromere probes, locus-specific probes, whole chromosome probes and telomere probes are some of the The whole chromosome probes are used for the multi-color FISH and spectral karyotyping. Take a look at the figure to know how different probes work. The pictorial illustration of the
Fluorescence in situ hybridization (FISH). FISH uses specific probes to determine the number of copies of a specific region of DNA that are present or whether a FISH utilizes fluorescently labeled probes that are complementary to and thus specifically hybridize a specific region of genomic DNA, allowing
FISH probes are typically derived from cloned genomic regions or flow-sorted chromosomes, which are labeled directly via nick translation or PCR in the Rather than relying on the isolation of a clone, such probes are designed to target precisely defined sequences. Also, as these probes are typically
Consequently, many gaps remain in our understanding of how local chromatin structure and nuclear positioning impact processes such as transcription, the sequences suitable to serve as an Oligopaint FISH probe. This density of variants allowed us to design 129-specic and. CAST-specic sets of

probe fish probes abl2 apart break

rna probe depiction representation mrna situ alkyne hybridization probes
Combine HCR IHC probes with HCR RNA-FISH probes to enable HCR protein-IHC + HCR RNA-FISH for multiplexed quantitative protein and RNA imaging with 1-step HCR Probe Design. Image any mRNA in any organism in any sample type. Straightforward multiplexing.

ros1 gene fusions identifying targeting aacrjournals clincancerres
Fluorescence in situ hybridization (FISH) is a molecular cytogenetic technique that uses fluorescent probes that bind to only particular parts of a nucleic acid sequence with a high degree of sequence complementarity.
Therefore, we have designed an interactive pipeline using the grid-based GLOBE 3D Genome Viewer and Platform to design and display different labelling variants of candidate probe sets. Thus, we have created a grid-based virtual "paper" tool for easy interactive calculation, analysis, management,
FISH-probe-design. For RNA or DNA FISH probes of any gene/transcript of interest. We use optional third-party analytics cookies to understand how you use so we can build better products. You can always update your selection by clicking Cookie Preferences at the bottom of
We designed oligo-FISH probes specific for one S. intermedia and one S. polyrhiza chromosome (Fig. 1a ). Our results show that these oligo-probes cross-hybridize with the homeologous regions of the other congeneric species, but are not suitable to uncover chromosomal homeology across

hybridization fluorescence procedure
FISH probes are composed of a fluorescent tag attached to a DNA fragment complementary to the DNA sequence being targeted. Control probes are used for both chromosome enumeration and verifying that copy number variation probes have hybridized to the correct chromosome.
This protocol describes the design of primer pairs against the human genome for the synthesis of probes for high-definition DNA FISH (HD-FISH). This pipeline selects PCR primer pairs with optimal thermodynamic features, delimiting amplicons 200-220 nucleotides in length, and filters out
To obtain specific FISH probes for interphase nuclei FISH studies of class switch recombination, we have developed and validated the webFISH algorithm by PCR primers were designed to generate PCR products for cloning into vectors used in FISH probe production. Six pairs of primers are for
4 Many commercial fish probes are available for common disorders or abnormalities… What happens for rare cases? 5 How to make your own probes Choosing DNA Growing and Knowing the sequence makes it possible to design a PCR reaction to test for its presence in any sample.

Can we use a specific primer or probe designed for Real time PCR as a FISH probe (off course with appropriate fluorophores)? The website has lots of information in regard to FISH probes and offers web-based and downloadable software for probe design and testing.
Design Guidelines for FISH Probes 1. Design multiple 24 to 30mer probes. Avoid stretches of more than 3 G or C bases. 2. To impart exonuclease resistance substitute 3-4 bases at the 5' and 3' end with 2'F bases.
fish- Fluorescence in situ hybridization. 1. Department of Biotechnology, Barkatullah University It can also be used to detect how many chromosomes of a certain type are present in each cell and to 4. Principles of fluorescence in situ hybridization (a) The basic elements of FISH are a DNA probe and
The 'FISH & Probes' section of the SILVA Webpage represents the current web-compendium for fluorescence in situ hybridization (FISH) of the Department for Molecular Ecology at the Max Planck Institute (MPI) for Marine Microbiology in Bremen, Germany.
Design FISH Probes. Use DECIPHER's Design Probes web tool to design the optimal FISH probes for targeting one group of RNA sequences in the presence of many non-target groups.
To design probes for multiplexed RNA FISH methods, ProbeDealer BLASTs oligos against the spliced transcriptome, and retain probes that only bind to For both chromatin tracing and RNA FISH probes, users may choose to output all probe sequences, or define how many probes they want for
Design Issue: Not Enough Probes. How to tell if you don't have enough probes Simply iterate your designs with varying probe lengths. Copy down the probe sequences recommended from each of these different length settings, and combine the non-overlapping probes into one set.
In this highly informative webinar, Daicel Arbor Biosciences will present the latest advancements in design and synthesis of custom FISH probes for use
How does FISH work?[ TOP ]. FISH is useful, for example, to help a researcher identify where a particular gene falls within an individual's chromosomes. Creative Biolabs offers a full array of custom fluorescence in situ hybridization (FISH) service from probe design, chromosome/cell preparation
7 How to Use the Fishbone Diagram? After that, you need to give bones to the fish. The causes are divided into six main branches in manufacturing, which are collectively referred to as the 6 Ms These will be designed to evaluate which of these potential reasons are actually contributing to the problem.
How are FISH probes used in DNA analysis? Apr 26, 2021 · To design probes for multiplexed RNA FISH methods, ProbeDealer BLASTs oligos against the spliced transcriptome, and Design your own Custom RNA FISH Probe Set to detect your RNA of interest. You can design up to 48
FISH tests using panels of gene-specific probes for somatic recurrent losses, gains, and FISH has also been used to detect infectious microbias and parasites like malaria in human blood cells. Fluorescence in situ hybridization (FISH) is a macromolecule recognition technology based on
Oligo FISH probes offer many advantages compared to conventional probes derived directly from genomic material. Synthetic oligos allow for (A) Diagram of the machine learning pipeline used to generate off-target scores for probe candidates. (B) Illustration of how dinucleotide counts
Hematology Probe Designs. The unique designs of the Vysis DNA FISH probes provide distinct advantages for detection of critical genetic aberrations associated The unique types of Vysis DNA FISH probe designs for chromosome translocations involved in these disorders are described below
