Learn more about DNA purity, how to measure it and how it affects quantification. Absorbance of DNA samples at 260 nm is currently most often measured using a microvolume spectrophotometer, but it is In this method, the concentration of a substance is calculated according to the
OF SIZE OF DNA Take the DNA of your interest and run it in 1% agarose gel. The DNA will move in the OF CONCENTRATION OF DNA
Tyrosine Kinase. DNA Damage/DNA Repair. The dilution calculator is a useful tool which allows you to calculate how to dilute a stock solution of known concentration. The dilution calculator is based on the following equation: Concentration(start) x Volume(start) = Concentration(final) x Volume(final).
DNA (deoxyribonucleic acid) concentration measurement is a commonly performed procedure in life DNA concentration was calculated based on the information provided in NIST SRM certificate of Following a similar approach, DNA concentration for an unknown sample can be
Calculations can be the bane of laboratory work. Fortunately, there are many easy methods to help you do the maths you need in the lab. Here, we tell you about the different ways to calculate primer concentration depending on the starting material. For all calculations, let's assume we have
21, 2020 · Here are Five DNA Quantification Methods to Consider UV absorbance. Using UV absorbance is one of the most common ways to quantify DNA. This method involves measuring Fluorescence dyes. Another way to quantify DNA would be to use fluorescent dyes that fluoresce when bound to DNA. ...
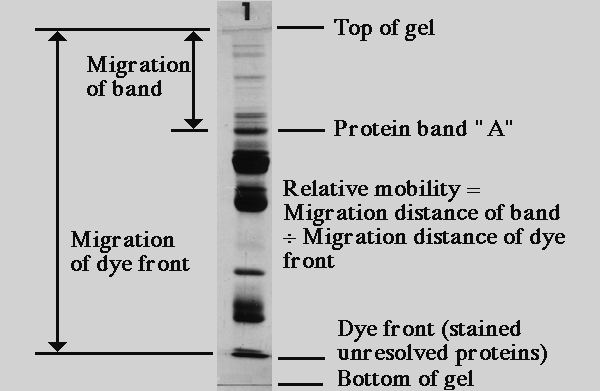
sds gel electrophoresis protein bands mobility polyacrylamide rf migration band relative measuring studies proteins mass calculator principle meaning apparent recipe
How to calculate the DNA concentration from A₂₆₀? How to compute the oligonucleotide sequence concentration? Average molecular weight of a nucleotide. How to calculate the extinction coefficients of DNA and RNA oligo sequences?
The DNA concentration calculated by multiplying it by 50 will be a BIG FAT LIE, that is, will be overestimated. So when there's an important protein contamination, the DNA concentration is overestimated. And how do I know if there's protein contamination? Easy: with clean
concentration is estimated by measuring the absorbance at 260nm, adjusting the A260 measurement for turbidity (measured by absorbance at 320nm), multiplying by the dilution factor, and using the relationship that an A260 of = 50µg/ml pure more on
Dna Concentration Calculation Economic! Analysis economic indicators including growth, development, DNA quantification methods. Economy. Details: Formula to calculate DNA concentration. dsDNA has an extinction coefficient of (µg/mL) -1 cm -1, hence: The
DNA concentration is estimated by measuring the absorbance at 260nm, adjusting the A260 measurement for turbidity (measured by absorbance at 320nm), multiplying by the dilution factor, and using the relationship that an A260 of = 50µg/ml pure dsDNA.
14, 2021 · What would be the molar concentration of human DNA in a human cell? Consult your teacher. The molar concentration of human DNA in a human diploid cell is as follows: ⇒ Total number of chromosomes × × 1023 ⇒ 46 × × 1023 ⇒ ×1018 Moles. What is molarity of DNA?
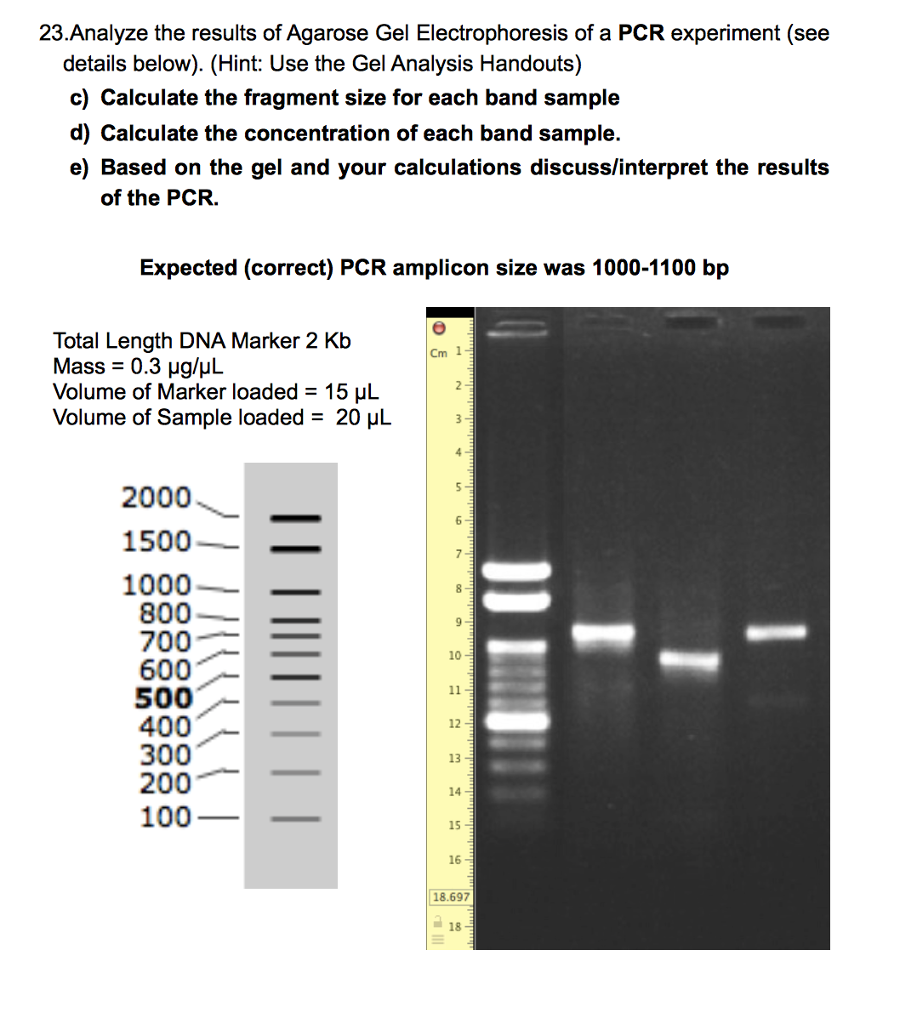
gel agarose results electrophoresis pcr analyze questions question chegg experiment answered transcribed hasn yet text been
The calculator calculates recommended Tm (melting temperature) of primers and PCR annealing temperature based on the primer pair sequence, primer concentration, and DNA polymerase used in PCR.
At lower concentrations, one cannot detect the DNA by sight or by noting the viscosity of the solution. When working with DNA, it is often The easiest means of determining DNA concentration is through spectrophotometric analysis. Since nitrogenous bases absorb UV light, the more concentrated
How do you calculate DNA concentration? Ad by Grammarly. DNA concentration is estimated by measuring the absorbance at 260nm, adjusting the A260 measurement for turbidity (measured by absorbance at 320nm), multiplying by the dilution factor, and using the relationship that an A260 of
How to calculate DNA concentration in Excel from. Details: This video explains about How to calculate DNA concentration in Excel from absorbance concentration can be from calculated molecular weight, ext dna concentration calc.

dna loading electrophoresis preparing gel flow dye load ppt powerpoint presentation underwater tip let slideserve

dna extraction proteinase method protocol methods buffer types different mentioned concentration required previous
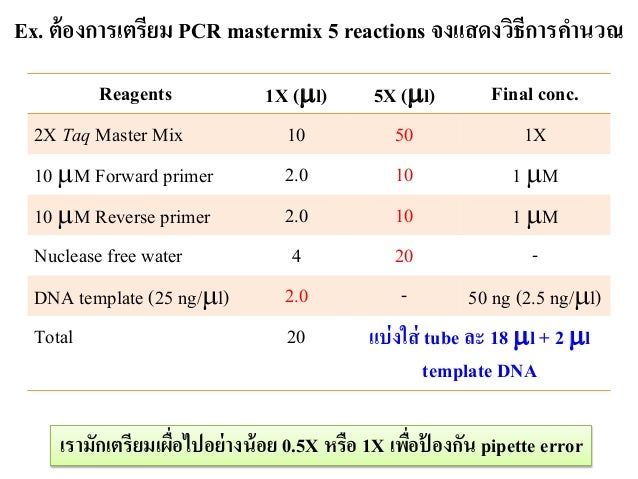
pcr mastermix ยม คำนวณ
How to calculate the DNA concentration and yield of DNA. Determining DNA Concentration (BIOL310)Подробнее. Molecular Diagnostics: Nucleic Acid Purity & Yield Calculation ExamplesПодробнее. Calculating DNA concentration and Experimental yieldПодробнее.
Calculate Nucleic Acid Concentration a. Divide the OD260 reading by the Table 3 pathlength value corresponding to the volume in the wells i. If a measurement volume not listed in Table 3 is used, calculate the pathlength correction using the linear regression equation shown in Table 3. b. Multiply this number by the DNA or RNA constant from Table 1
Calculate nucleic acid concentration from absorbance and vice versa. Calculate base-pair molarity of dsDNA solutions. For DNA and RNA oligonucleotides, the calculations based on predicted extinction coefficients give more accurate results.
› Get more: How to quantify dnaShow All. How to calculate DNA concentration? AAT Bioquest. How. Details: The DNA concentration can be determined by measuring the absorbance of the sample at 260nm in spectrophotometer and use the Beer-Lambert's law to calculate the concentration.
For the measurement of the concentration of nucleic acids (DNA , RNA) different A prototype fluorometer has been built (Images shown below) for measuring the concentration of nucleic acids. The V - I characteristic is represented in the graphic below in which we see how the curve, in

transcribed
UV for quantification of nucleic acid concentration. § Is determined by measuring absorbance at of the DNA and to calculate the sizes. the use of appropriate size markers. To see if your DNA How you will choose the appropriate concentration of the gel ? § What are the things that you
fludarabine phosphate stat1 synthesis activity apexbt
can u help me ,how to calculate the concentration???? we have a standard picture of gel electorphoresis of serial dilution of DNA it starts from 5ng, 10 ng, 50 ng, 100 ng, 200 ng, 500ng, 1 ug etc.
Precise assessment of DNA concentration is of crucial importance in many analytical processes and medical diagnostic applications. Recently, we found that hydrodynamic forces can lead to DNA motion against the electrophoretic force (EPF) at low ionic strength.
May 05, 2020 · How do you calculate DNA concentration from od260? To determine the concentration of DNA in the original sample, perform the following calculation: dsDNA concentration = 50 μg/mL × OD260 × dilution factor. dsDNA concentration = 50 μg/mL × ×
3. Calculate DNA concentration and from agarose gel electrophoresis results 4. Calculate DNA concentration from UV absorbance results. 6. Information on how to use the Beckman 500 UV spectrophotometer are given below, but take readings as directed by your instructor.
08, 2017 · Just use undiluted stock DNA in these cases. // For each stock concentration (sampleConc): // Rearranged c1 * v1 = c2 * c2 var rawTemplateVol = (finalVol * finalConc) / sampleConc; // If DNA volume for desired concentration is < minimum volume, find the scaling-up factor. // If DNA volume is not less than the minimum volume, then the scaling factor is 1. …
08, 2021 · To find the 260/280 ratio, proceed as follows: Measure the absorbance of the sample at the wavelength of 260 nm (A₂₆₀). Measure the absorbance of the sample at the wavelength of 280 nm (A₂₈₀). Divide A₂₆₀ by A₂₈₀ to obtain the ratio. Author: Michael Darcy
This video explains about How to calculate DNA concentration in Excel from absorbance concentration can be from calculated molecular
This protocol describes how to run a standard agarose gel utilizing concentration and size standards as well as Qubit™ fluorometer to evaluate the quality Cast a ~100ml 1% agarose gel with 1X TAE and ethidium bromide (.15ug/ml) or SYBR® Safe DNA gel stain (10,000X concentrate in DMSO).
Calculating the DNA copy number is an important step in absolute qPCR since the number of copies are required Use our DNA copy number online calculator. Are you still struggling with the maths? If so, I have also created an If you would like to calculate the concentration in pg/mL, you can

excel concentration unknown sample determining
Fortunately, understanding how base pairs function and even calculating percentages for each base in a DNA sample is actually straightforward. They total 100 percent of the sample. Chargaff's rule states that the concentration for each base in a base pair is always equal to its mate, so the
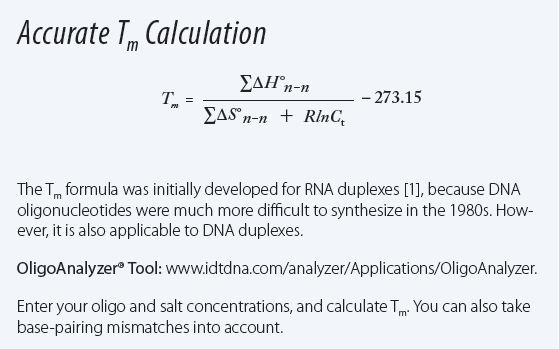
tm melting temperature formula salt temp inset understanding environment decoded yr d3
DNA conc. is calculated by this aquation; OD of diluted dsDNA at 260nm * Dilution factor ( 10ul of extracted DNA/990ul of high pure water=100) * 50 With traditional UV spectrophotometer, you will need a DNA standard with know concentration. You will also need a fluorescent dye (eg. pico-green).
How do you know if you actually have DNA in your tube without seeing it? There are many ways to do this and the method you choose could be based on your downstream application, time, and instrument availability. Then, using the A260 reading, you can calculate the DNA concentration.
05, 2020 · To determine the concentration of DNA in the original sample, perform the following calculation: dsDNA concentration = 50 μg/mL × OD260 × dilution factor. dsDNA concentration = 50 μg/mL × × 50. dsDNA concentration = mg/mL.
31, 2018 · Nucleic Acid Concentration. DNA Concentration Calculator. Answer. The DNA concentration can be determined by measuring the absorbance of the sample at 260nm in spectrophotometer and use the Beer-Lambert's law to calculate the concentration. In order to do so, you would need to determine the extinction coefficient of the DNA given in cm-1M-1.
» OD260 Nucleotide Concentration Calculator. Note: The nucleic acids (DNA, RNA and oligo) solutions with different concentration has different ability aborbing light. When the light wave is 260 nm, the Absorbance of light and the nucleic acid concentration is calculated as: C = A/(e * l)
~mbumbuli/molbio/labs/dnaSince nitrogenous bases absorb UV light, the more concentrated the DNA solution, the more UV light it will absorb. Rule: The concentration of pure double-stranded DNA with an A260 of is 50 mg/ml. Thus, one can use the following formula to determine the DNA concentration of a solution. Unknown (mg/ml)/ Measured A260 = 50 (mg/ml)/ A260
Two main protocols exist for determining DNA concentration, depending on whether you want to be able to measure samples over a Broad Range (BR) or with a high High Sensitivity (HR) for lower DNA protocols are identical, but require the use of different buffer, dye, and standards.
